The Decorated RNA Supermolecular World and A Method to Develop Novel Ligands and Catalysts
By Tianxin Wang
In recent years, continuous
discoveries of catalytic RNA and DNA by the in vitro selection method greatly
support the RNA World theory[1], which assumes that the chemical
process leading to the appearance of life was carried out by RNA. Trying to
overcome the structural limitation of the four bases, ingenious strategies such
as introduction of coenzymes and unnatural bases [2 ]
have been developed. However, it seems that the ultimate catalytic
potential of nucleic acid has yet to be fully exploited. Does this imply that
the structural types of these ribozymes generated by in vitro selection are only a small
portion of the vast existing ribozymes in the RNA (DNA) World? In the RNA
library for in vitro selection, the RNAs are generally
single-strained ¡°pure¡± nucleic acid in similar length. This is unlikely to occur
in the pristine world. These
postulations raise an interesting question:
Must the ribozyme in the RNA World be a
single-strained ¡°pure¡± RNA? Could it be a supermolecular system consisting of
several partially complementary RNA strands, among which, some would be
covalently conjugated
(modified) by other
molecules?
Orgel¡¯s report
on the genetic take over
[3] of nucleic acid shows that many short complementary nucleic acids
of different length have been generated by a long nucleic acid template during
its self replication. This experiment suggests that in the primitive soup of
nucleic acids, there were not only many long nucleic acids but also more short
nucleic acids which are complementary to these long nucleic acids since at that
time the fully developed RNA polymerase had not evolved yet. Thus the generation
of nucleic acid supermolecular systems (Fig.1) should be a general phenomenon.
Therefore in the primitive world, some enzyme-like nucleic acids might be single
stranded RNA while other such nucleic acids might be supermolecular systems
which are consisted of several partially complementary RNA strands, just as some
protein enzymes are consisted of several subunits.
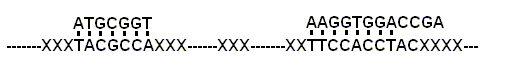
Figure 1. Self-assemble RNA
Supermolecular System (x: varied nucleotides)
The advantage of
the self-assemble strategy is that sophisticate structures and higher diversity
can be generated from fewer and smaller building blocks, a strategy similar to
that antibody employs for more functions. The tradeoff is that lower rigidity of
the structure may cause lower efficacy, therefore building blocks other than
nucleotides are necessary to compensate the efficacy.
Since the primitive soup
contains both amino acids and nucleic acids, there should be some chemical
connections between these two main kinds of bio-molecules. First, research
[4] shows that phosphoryl amino acids can generate short peptide
decorated nucleic acids in mild environment, indicating that some chemical
activators might generate the hybridization of peptides and nucleic acids in
primitive world. Secondly, the discovery of amino acid transfer
ribozyme[5] reveals the possibility of the existence of nucleic
acid-amino acid conjugation (Fig.2). Since more and more ribozymes
that catalyze alkylation, acylation, cycloaddition reactions on the RNA
substrate [6] producing RNA-other molecule conjugation have been
discovered, we expect that some other small molecules decorated RNAs (Fig.3)
could also exist in the primitive world. Some long RNAs from these conjugations
may have new catalytic functions, because introducing new function groups would
overcome nucleotides¡¯ structural limitation and give propensity of generating
novel activity.
Fig.2 RNA -amino acid
conjugation
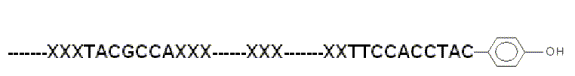
Fig.3 RNA-small organic
molecule conjugation
Currently most
artificial ribozymes¡¯ catalytic activities are self-modification. An enzyme
which can self modify and generate other catalytic function thereafter is rare.
In the primitive world, a probable strategy might be that: one group of
ribozymes modify short, partially complementary nucleic acids with amino acid
(Fig.2) or other small organic molecules. The modified short nucleic acids then
self assemble with other long nucleic acids to form supermolecular systems
(Fig.4). Some of these
supermolecular systems would have ability to catalyze special reactions. This
postulation could be supported by (or explain) the fact that most current
catalytic RNAs need a RNA fragment as substrate.
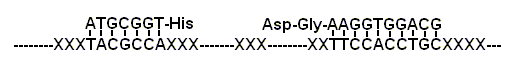
Figure 4 An amino acid decorated RNA system, which can be regarded as a RNA that can utilize RNA coded amino acids cofactors
If life did originate from
the RNA world, a mixed and modified RNA world would be more plausible than a
single strand, pure RNA world. In
fact, the first discovered ribozyme is also a supermolecular system consisting
of not only RNA, but also Mg2+ and guanosine. Recent discovery that
the RNA-RNA interaction can be an activator for RNA synthesis shows us again the
power of complementary base pairing [7]. It is reasonable to assume
that nature would use such supermolecule to build more sophisticated ribozymes,
since it is such a wonderful tool for biofunctions.
Along the evolution course,
the amino acids and peptides on the nucleic acid backbone would become longer
and longer while the nucleic acid parts would become shorter and shorter and
eventually vanish completely. Then the evolution from ribozyme to protein enzyme
would complete. Therefore, we may postulate that some of the modern intron could
be the relic of these complementary nucleic acids fragments, and that the
peptide modified RNA (DNA) would be the missed link between the RNA (DNA) enzyme
and protein enzyme.
A catalytic DNA using amino
acid as cofactor has been discovered recently [2 ].
It is more convenient that ribozymes utilize DNA coded cofactors (Fig. 5) rather than a
cofactor alone to expand
their catalytic potential. Today, there are still many nucleotide-based
coenzymes such as NADH and other conjugation such as tRNA, which may be the
relic of these kinds of original RNA (DNA) cofactor
conjugations.
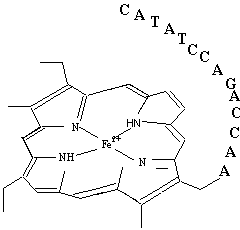
Fig.5 A DNA-porphyrin conjugation as a DNA coded cofactor
In in vitro selection for new catalysts and
useful ligands, DNA coded cofactor
may also help. For a RNA (DNA) library such as:
----XXXXXXXXATTGGCTTCGXXXXXXXXTATACCCXXX---,
we can add
chemically synthesized complementary RNA, DNA, or PNA (peptide nucleic acid)
fragments such as TAAXXXXCCGAA-Gly,
ATATGGG-Ala, and ATATGGG-His
into the system to form a supermolecular library. The supermolecular structure
may have superior affinity or catalytic activity due to the amino acid parts.
There are infinite new
structures such as unnatural small molecules, chelate-metallic ions and
coenzymes, and large systems such as C60 and nanoparticles even
molecular devices. They can be linked to nucleic acid fragments by organic
synthesis easily. By introducing these new building blocks into in vitro selection system (Fig.6), we
may be able to do better than nature to create more novel
functions.
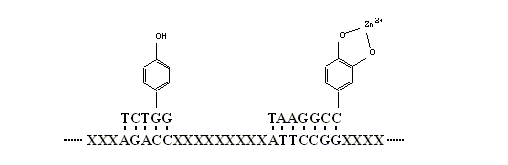
Figure 6 A decorated DNA
supermolecular library containing novel functional groups for new
activities
Reference:
1) Walter Gilbert: Nature, 1986, Vol.319,
618
2) Kandasamy Sakthivel,
Carlos F. Barbas: Angew. Chem .Int.
Ed. 1998 (37), 2872
Adam Roth, Ronald R.
Breaker: Proc. Natl. Acad. Sci. USA
1998 (95)
6027-6031
T.M.Tarasow, S.L.Tarasow,
B.E.Eaton: Nature, 1997,389,54-57
3) Christof Bohler, Peter E.
Nielsen, Leslie E. Orgel: Nature, 1995, 376,
578-581;
4) Zhao Yufeng, Li Yanmei:
J. of Biochemical Physics, 1994, 20, 283-287; Chinese Chemical Letter, 1996, Vol 7. No 10, 905-906,
5) Lohse P.A, Szostak J.W:
Nature, 1996, 381,
442-444
6) Peter J. Unrau, David P.
Bartel Nature 1998, 395,
260-263;
Wilson C, Szostak J.W: Nature, 1995, 374 ,777-783;
Illangasekare M, Sanches G,
Nickles T, Yarus M, Science, 1995,267,643-647
7) Sit T.L, Vaewhongs A.A, Lommel S.A. Science 1998 , 281, 829-832